Mutagenesis and lateral gene transfer: processes implicated in the bacterial defense against bacteriophages
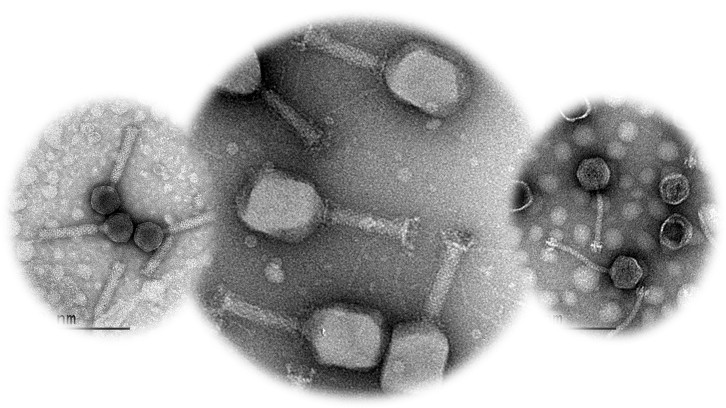
One of the major health challenges nowadays is the fight against bacterial resistance to antibiotics. The use of bacteriophages (virus that only infect bacteria) is one of the most promising strategies for that purpose, but bacteria can also develop resistance and defense mechanisms against them. The Molecular Microbiology group of the Department of Genetics and Microbiology studies how these resistance and defense mechanisms emerge and has obtained outstanding results in a work where a three-bacteriophage cocktail is used against Salmonella enterica serovar Typhimurium, one of the main agents that cause gastrointestinal disease in humans.
One of the main health problems of the 21st century is the bacterial resistance to antibiotics, and the development of new strategies to overcome it globally from a One Health perspective is a pressing matter. Phage therapy has gained special relevance to fight this alarming reality. However, similarly to what has happened with the emergence of resistance to antibiotics, the use of phages in pathogen control can result in the emergence of bacterial variants resistant to phages.
The Molecular Microbiology group of the Department of Genetics and Microbiology of the UAB, led by Dr. Montserrat Llagostera, has studied the emergence of resistance and defense mechanisms to a three-phage cocktail against the pathogen Salmonella enterica serovar Typhimurium. Using conventional and molecular microbiology methods and bioinformatics, several Salmonella variants with reduced susceptibility to the phages were isolated from laboratory cultures of Salmonella infected with phages (LAB), from cooked ham slices contaminated with Salmonella and treated with the phages as a model of food security (FOOD), and from oral phage therapy in broilers contaminated with Salmonella (PT). This study, published in Frontiers in Cellular and Infectious Microbiology, demonstrates that the isolation frequency of these variants in LAB is much higher than in FOOD and PT.
It has been demonstrated that the causes of the emergence of these variants are different in each scenario. In LAB and FOOD, these variants are resistant to the phages thanks to mutations in the genes rfc and rfaJ involved in the synthesis of the phage receptors. However, no variants with resistance to the phages were detected in PT. Instead, these variants have a mechanism of interference with the multiplicative cycle of the phages. These mechanisms are found in conjugative plasmids present in the intestinal microbiota of the broilers. These results prove for the first time that plasmid acquisition through lateral transfer can have more relevance than mutagenesis and that the probability of these events happening doesn’t significantly affect the success of food biocontrol and oral phage therapy.
M. Pilar Cortés Garmendia
Department of Genetics and Microbiology
Universitat Autònoma de Barcelona
mariapilar.cortes@uab.cat
Montserrat Llagostera Casas
Department of Genetics and Microbiology
Universitat Autònoma de Barcelona
montserrat.llagostera@uab.cat
References
López-Pérez J, Otero J, Sánchez-Osuna M, Erill I, Cortés P, Llagostera M. Impact of mutagenesis and lateral gene transfer processes in bacterial susceptibility to phage in food biocontrol and phage therapy. Front. Cell. Infect. Microbiol. (2023). Volume 13 - 2023 | doi: 10.3389/fcimb.2023.1266685


